Maulik Kamdar | Elsevier's Healthcare Knowledge Graph
KGC | All Access Subscription
•
20m
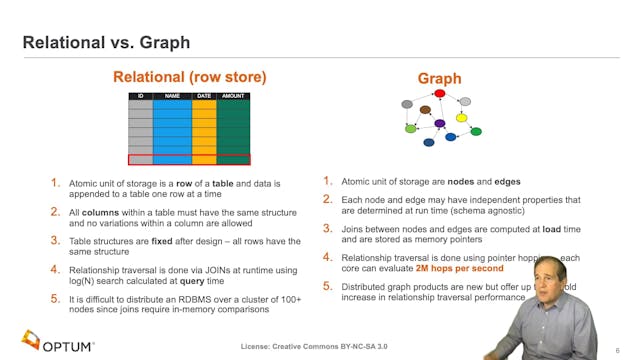
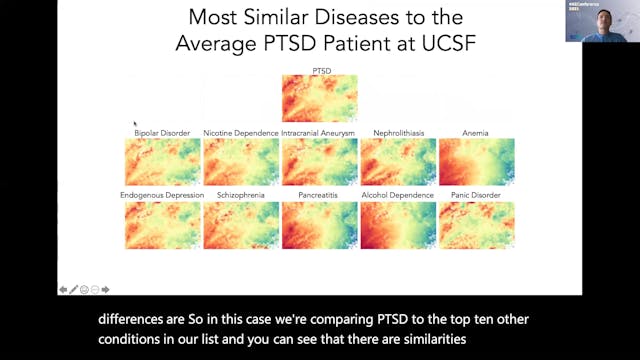
Knowledge Graphs are increasingly being developed and leveraged in academia and industry to tackle complex biomedical challenges, such as drug discovery and safety, medical literature search, clinical decision support, and disease monitoring and management. In this talk, we will present the research and development on Elsevier’s Healthcare Knowledge Graph, a platform built to deliver advanced clinical decision support and enhanced point-of-care content discovery for clinicians and patients. Elsevier’s Healthcare Knowledge Graph uses popular linked data and semantic web technologies to capture knowledge and data from heterogeneous healthcare sources about diseases, drugs, findings, guidelines, cohorts, journals, and books. The graph is composed of medical concepts (e.g., drugs, symptoms), medical term labels and synonyms for these concepts, hierarchical and associative typed relations (e.g. drug—disease relations) between these concepts, and mappings to codes in external terminologies used in clinical data integration and electronic medical record systems. We will demonstrate how subject matter experts curate and visualize the Healthcare Knowledge Graph through novel exploration interfaces to keep the medical knowledge regularly updated, and also showcase how operationalized natural language processing pipelines continuously tag and extract novel medical concepts and relations within synoptic and reference medical content. We will walkthrough how Elsevier’s Healthcare Knowledge Graph platform is used to provide actionable knowledge in user-facing applications in the domains of focused and precise clinical search, clinical decision support, and content recommendation. This talk will provide a perspective on how such knowledge graphs will enable the capture, representation, and provision of complex medical knowledge of high velocity, variety, volume, and veracity, to power trusted clinical and biomedical research applications.
Up Next in KGC | All Access Subscription
-
Dan McCreary | Graph Hardware Is Coming!
In this presentation we will show how current general-purpose CPU hardware fails to deliver high performance graph analytics. We show that by doing a detailed analysis of the actual hardware functionally needed by graph queries (pointer jumping), we can redesign hardware that is optimized for fas...
-
Olaf Hartig | RDF Star: Metadata For ...
The lack of a convenient way to capture annotations and statements about individual RDF triples has been a long standing issue for RDF. Such annotations are a native feature in other contemporary graph data models (e.g., edge properties in the Property Graph model). In recent years, the RDF* app...
-
Sergio Baranzini | SPOKE: A Biomedica...
SPOKE is a biomedical knowledge graph containing factual data on various biomedical fields including genetics, molecular biology, physiology, metabolomics, pharmacology and clinical medicine. SPOKE can be used to repurpose medications, predict patient outcomes, and accelerate drug development, am...